操作步骤
- 使用PuTTY工具,以root用户登录服务器。
- 执行以下命令下载算例文件。
wget http://labshare.cshl.edu/shares/schatzlab/www-data/ectools/w303/Illumina_500bp_2x300_R1.fastq.gz wget http://labshare.cshl.edu/shares/schatzlab/www-data/ectools/w303/Pacbio.fasta.gz
- 执行以下命令解压算例文件。
gzip Pacbio.fasta.gz –d gzip Illumina_500bp_2x300_R1.fastq.gz -d
- 执行以下命令处理数据。
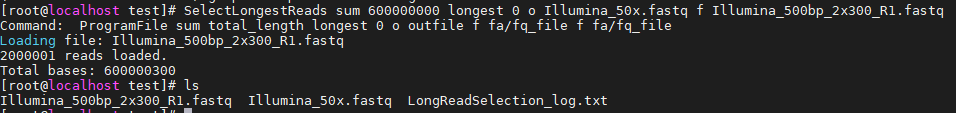
SelectLongestReads sum 600000000 longest 0 o Illumina_50x.fastq f Illumina_500bp_2x300_R1.fastq
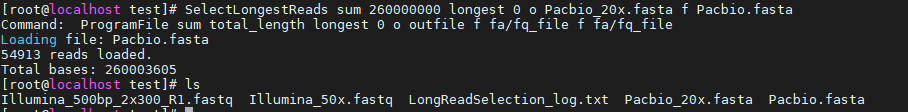
SelectLongestReads sum 260000000 longest 0 o Pacbio_20x.fasta f Pacbio.fasta
- 执行以下命令创建“Illumina_data”目录,并将生成的fastq文件拷贝至“Illumina_data”目录中。
mkdir Illumina_data && cp Illumina_50x.fastq Illumina_data/
- 执行以下命令创建“Pacbio_data”目录,将生成的fasta文件拷贝至“Pacbio_data”目录中。
mkdir Pacbio_data && cp Pacbio_20x.fasta Pacbio_data/
- 执行以下命令创建并进入“step1”目录。
mkdir step1 && cd step1
- 执行以下命令利用Illumina小片段文库数据组装出contigs序列。
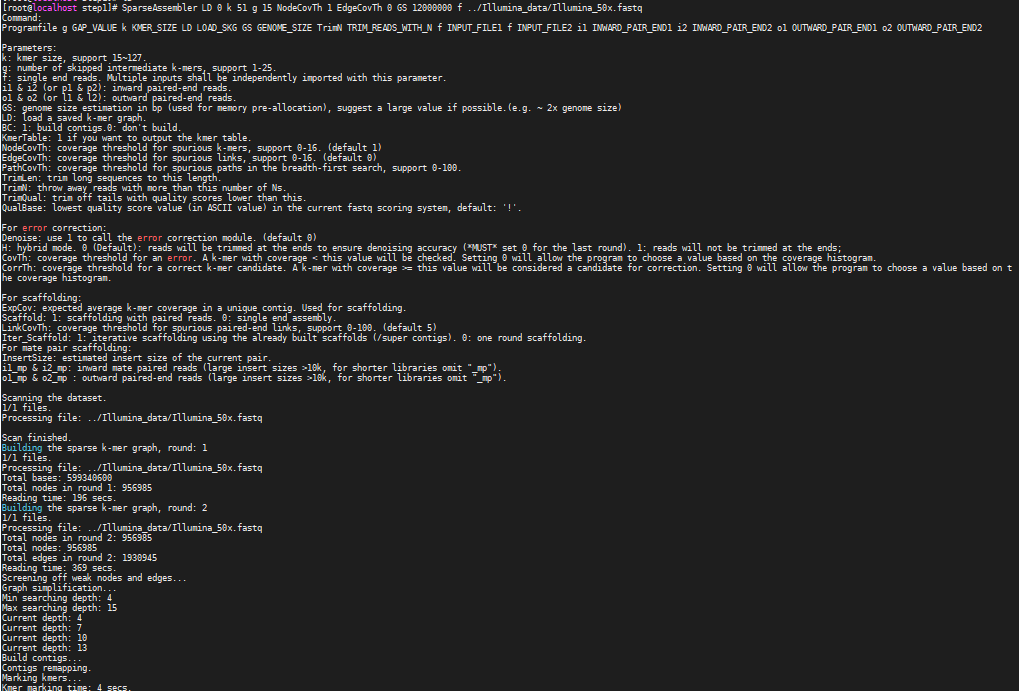
SparseAssembler LD 0 k 51 g 15 NodeCovTh 1 EdgeCovTh 0 GS 12000000 f ../Illumina_data/Illumina_50x.fastq
SparseAssembler LD 1 NodeCovTh 2 EdgeCovTh 1 k 51 g 15 GS 12000000 f ../Illumina_data/Illumina_50x.fastq
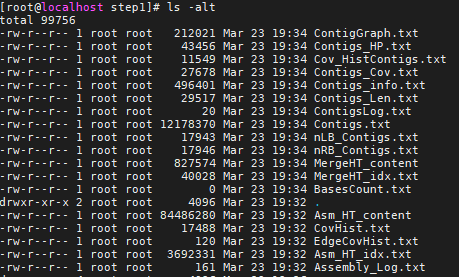
生成如下文件。
- 执行以下命令找Contigs序列和Pacbio reads的Overlap并进行Layout。
DBG2OLC k 17 AdaptiveTh 0.0001 KmerCovTh 2 MinOverlap 20 RemoveChimera 1 Contigs Contigs.txt f ../Pacbio_data/Pacbio_20x.fasta
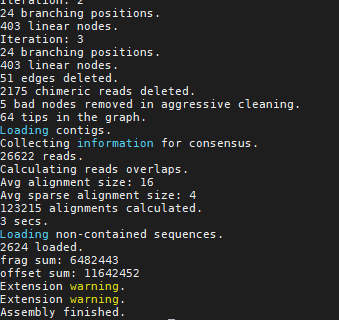
回显显示信息如下图所示。
生成如下文件。

- 使用“/opt/biosoft/DBG2OLC/utility/”目录下的“python”和“shell”脚本,调用blasr和consensus模块的Sparc进行运算。
- 执行以下命令修改“split_and_run_sparc.sh”脚本。
vi split_and_run_sparc.sh
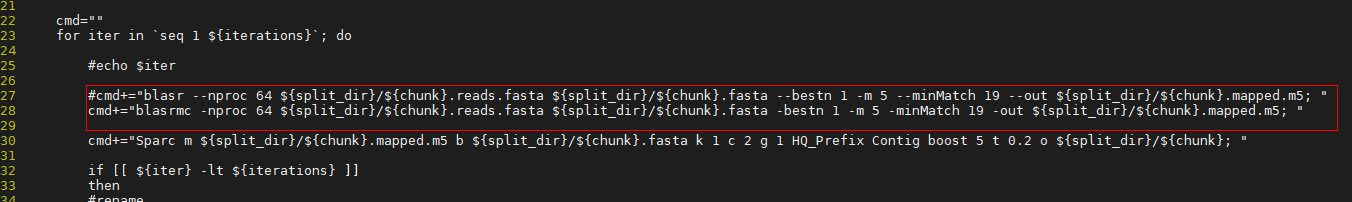
- 按“i”进入编辑模式,注释第27行的内容,添加第28行的内容。
- 按“Esc”键,输入:wq!,按“Enter”保存并退出编辑。
- 在step1目录下执行以下命令。
cp ../Pacbio_20x.fasta . cat Contigs.txt Pacbio_20x.fasta > ctg_pb.fasta mkdir consensus_dir split_and_run_sparc.sh backbone_raw.fasta DBG2OLC_Consensus_info.txt ctg_pb.fasta ./consensus_dir 2 >cns_log.txt
生成如下文件。
- 执行以下命令修改“split_and_run_sparc.sh”脚本。